Computational Data Science
My activities within those of the
ADAPT (Advanced Data Analysis and Parallel computing Team) group
(formerly CDS-group and
CDS-Lab), ICAR-CNR
Objectives
The activity concerns the design of models, algorithms, and software tools to discover, understand, and model scientific phenomena through the analysis of experimental data and/or through the simulation of their generation processes.Main achievements
Omics Imaging See [29], [31] for an overview.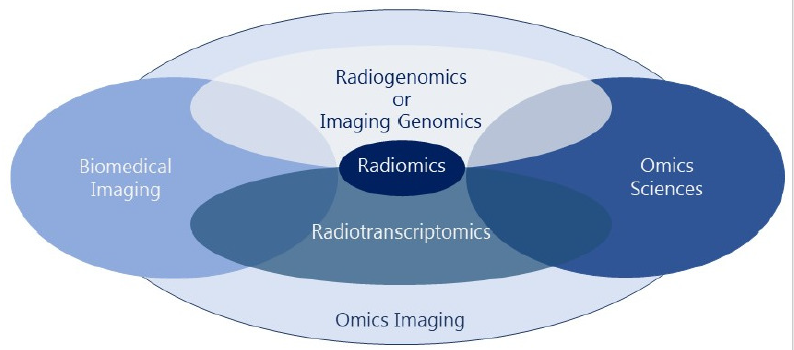 - Integrating Imaging and Omics Data [1],[4]. See also our companion web page on OmicsImaging.
- Integrating Imaging and Omics Data [1],[4]. See also our companion web page on OmicsImaging. |
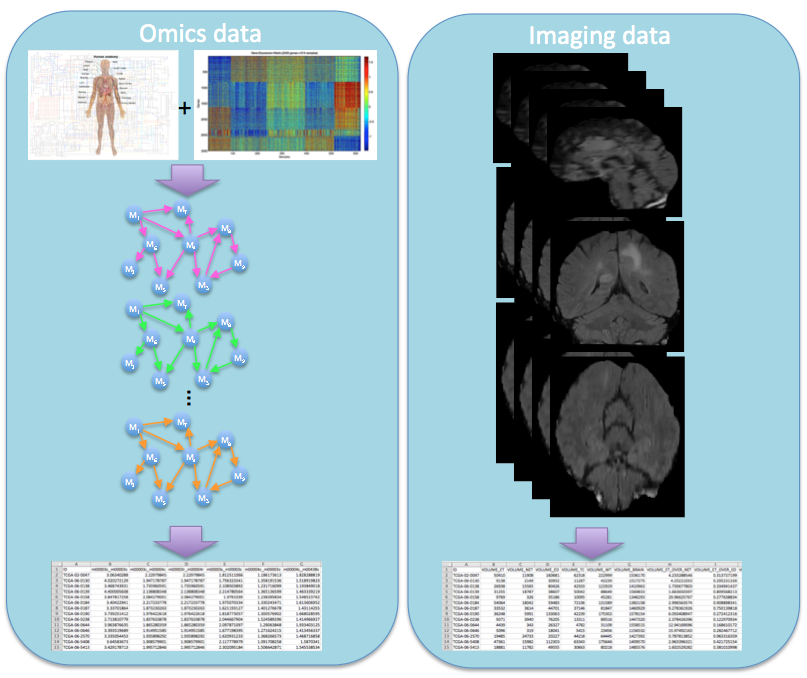 - Glioma Grade Classification via Omics Imaging [6],[10],
Best Paper Award at the 7th International Conference on Bioimaging (BIOIMAGING2020).
- Glioma Grade Classification via Omics Imaging [6],[10],
Best Paper Award at the 7th International Conference on Bioimaging (BIOIMAGING2020).
|
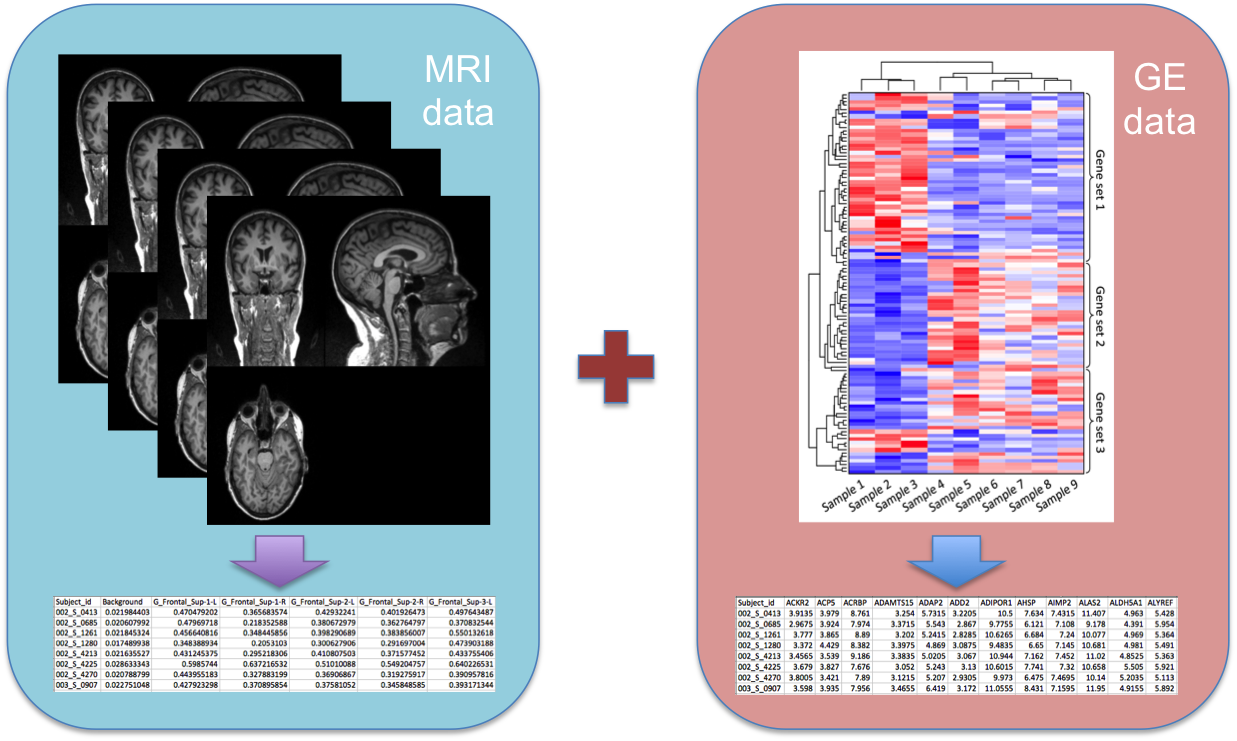 - Classification and Clustering of Alzheimer's Disease Using MRIs and Transcriptomic Data [20],[25], [34].
- Classification and Clustering of Alzheimer's Disease Using MRIs and Transcriptomic Data [20],[25], [34].
|
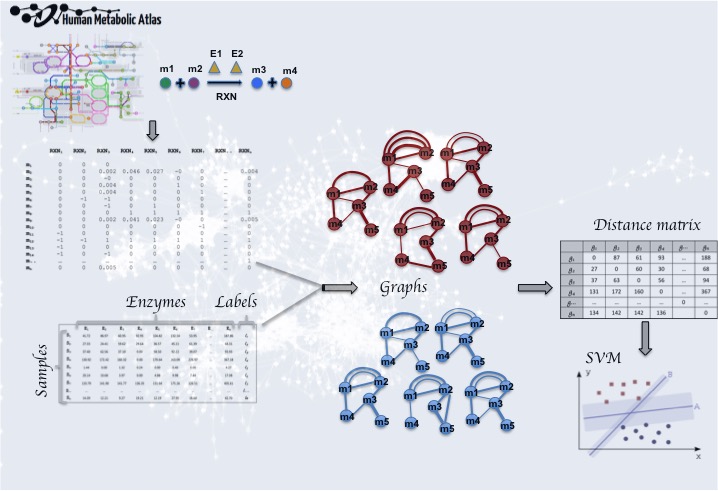 - Clustering [2],[8] and classification [3],[5],[7] of metabolic networks [22].
See also our poster on Supervised and Unsupervised Learning on Biological Networks (2019).
- Network similarities [7],[9].
- Clustering [2],[8] and classification [3],[5],[7] of metabolic networks [22].
See also our poster on Supervised and Unsupervised Learning on Biological Networks (2019).
- Network similarities [7],[9].
| 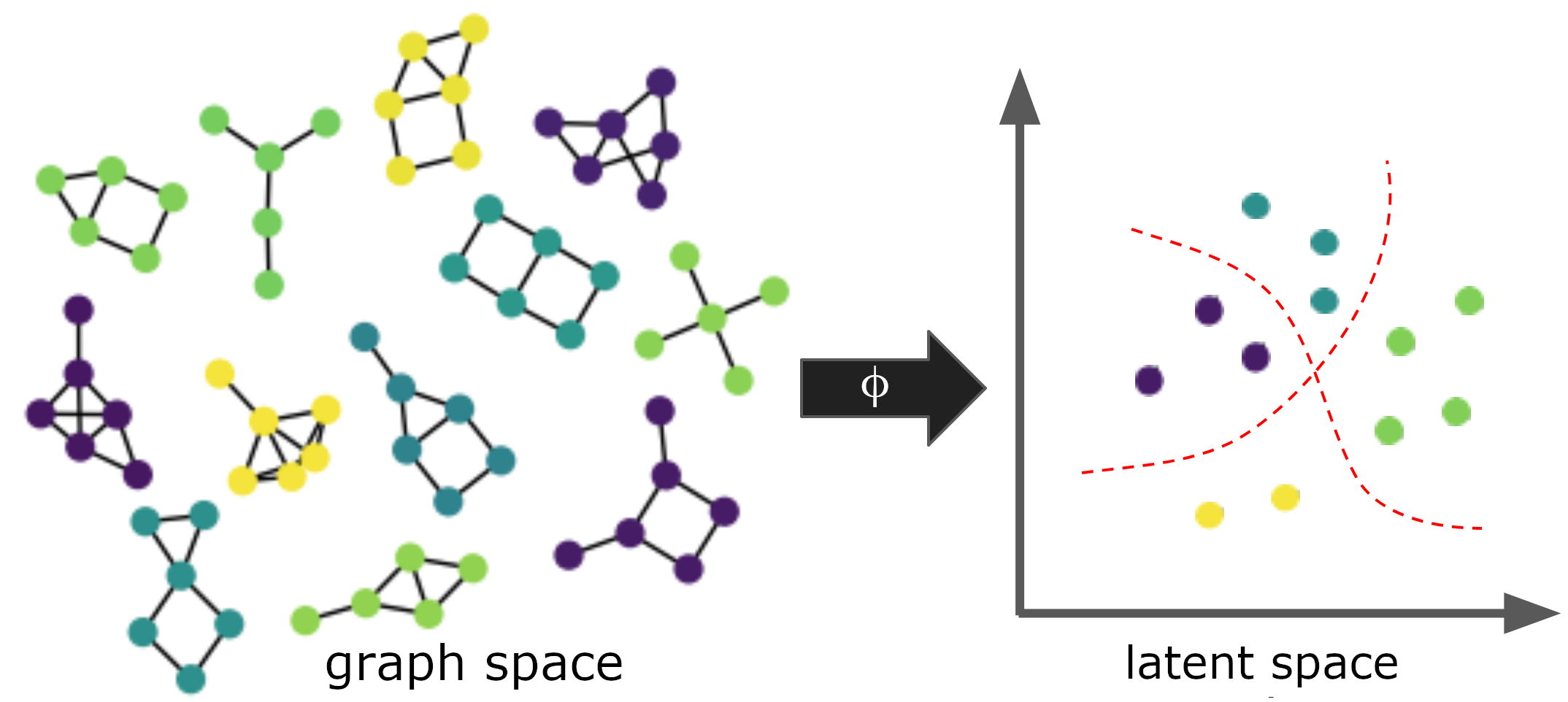 - Whole-graph embedding [11],[12],[15],[19]
and its robustness [13],[14],[16],[17],[18],[21],[23].
- Community detection [24].
- Gene essentiality identification [26], [28], [30],
[32], [33], [35].
- Graph Neural Networks [36], [37].
- Whole-graph embedding [11],[12],[15],[19]
and its robustness [13],[14],[16],[17],[18],[21],[23].
- Community detection [24].
- Gene essentiality identification [26], [28], [30],
[32], [33], [35].
- Graph Neural Networks [36], [37].
|
Main awards
- Challenge Winners: The CDS-Lab Team (Mario Rosario Guarracino, Maurizio Giordano, Ichcha Manipur, Ilaria Granata, Lucia Maddalena, Marina Piccirillo) is among the winners of the first subchallenge of the Metagenomics Diagnosis for IBD Challenge (MEDIC), September 2020 [27]. - Best Paper Award: 7th International Conference on Bioimaging (BIOIMAGING/BIOSTEC) for Glioma Grade Classification via Omics Imaging, by L. Maddalena, I. Granata, I. Manipur, M. Manzo, and M.R. Guarracino, February 26, 2020.Main collaborations
- National Research University Higher School of Economics, Lab LATNA, Russia - Stazione Zoologica “Anton Dohrn”, Naples, Italy - University of Cassino and Southern Lazio, Italy - University of Florida, U.S.A. - University of Naples "L'Orientale", Naples, ItalyUseful/downloadable material
Dataset - TumorMet: repository of tumor metabolic networks derived from context-specific Genome-Scale Metabolic Models, presented in [22] Software - MetabolitesGraphs: R package to integrate gene expression data into a metabolic model and extract metabolites-based graphs, as used in [3],[5],[9] - GraphDistances: R package to compute distribution-based distance measures between directed and undirected graphs/networks, as used in [3],[5],[9] - Netpro2vec: Python code for the neural embedding framework proposed in [11] - OI_Glioma: Matlab code and data for the Glioma Grades classification framework proposed in [10]. - OI_AD: Matlab code and data for the Alzheimer's Disease classification framework proposed in [25]. - EG-identification: Python code and data for reproducing the experimental results of [26]. - HELP [33]: computational framework for labelling and predicting essential genes in different human tissue and disease contexts described in [32]. Web pages - OmicsImaging: Compendium of [4]Publications and Communications on Computational Data Science
[37] B.D. Bernhardt, C. Marciano S. Gigli, L. Maddalena, and M.R. Guarracino, Understanding ESG Scores Through Network Analysis: A Study Using Graph Neural Networks, in A. D'Ambrosio, M. de Rooij, K. De Roover, C. Iorio, M. La Rocca (eds) Supervised and Unsupervised Statistical Data Analysis. CLADAG-VOC 2025. Studies in Classification, Data Analysis, and Knowledge Organization. Springer, Cham., DOI: 10.1007/978-3-032-03042-9_4, 2025. [36] C. Marciano, B.D. Bernhardt, L. Maddalena, and M.R. Guarracino, EGNAN: An Enhanced Method for Interpretable Graph Neural Networks, in E. di Bella, V. Gioia, C. Lagazio, S. Zaccarin, S. (eds) Statistics for Innovation III. SIS 2025. Italian Statistical Society Series on Advances in Statistics. Springer, Cham., DOI: 10.1007/978-3-031-95995-0_76, 2025. [35] M. Giordano, L. Maddalena, M. Manzo, M.R. Guarracino, I. Granata, Context-specific Essential Genes Identification and Prediction by Learning Multi-Omics and Network Data, Oral presentation at Bioinformatics and Computational Biology Conference 2024 (BBCC2024), Naples, November 27-29, 2024. [34] L. Antonelli, C. Di Napoli, L. Maddalena, G. Paragliola, P. Ribino, L. Serino, I. Granata, Clustering Based Stratification of Mild Cognitive Impairment: Insights from Blood Transcriptomic Data, Poster at Bioinformatics and Computational Biology Conference 2024 (BBCC2024), Naples, November 27-29, 2024. [33] M. Giordano, I. Granata, L. Maddalena, HELP: Human Gene Essentiality Labelling & Prediction, computational framework for labelling and predicting essential genes in different human tissue and disease contexts. DOI: 10.5281/zenodo.12697605, 2024. [32] I. Granata, L. Maddalena, M. Manzo, M.R. Guarracino, M. Giordano, HELP: A computational framework for labelling and predicting human common and context-specific essential genes, PLoS Computational Biology 20(9), DOI: 1371/journal.pcbi.1012076, 2024. Software and data available here and here. [31] L. Maddalena, Computational Data Science Approaches for Biomedical Data, communication at CSMM2024: Calcolo Scientifico e Modelli Matematici - Alla Ricerca delle Cose Nascoste Attraverso le Cose Manifeste, Naples, January 30, 2024. [30] M. Giordano, E. Falbo, L. Maddalena, M. Piccirillo, I. Granata, Untangling the Context-Specificity of Essential Genes by Means of Machine Learning: A Constructive Experience, Biomolecules 14(1), ISSN: 2218-273X, DOI: 10.3390/biom14010018, 2024. Data available here. [29] L. Maddalena, Some Experiences with Omics Imaging, Keynote talk at 23rd International Symposium on Mathematical and Computational Biology (BIOMAT 2023), November 2023. [28] I. Granata, M. Giordano, L. Maddalena, M. Manzo, M.R. Guarracino, Network-Based Computational Modeling to Unravel Gene Essentiality, in R.P. Mondaini (Ed), Trends in Biomathematics: Modeling Epidemiological, Neuronal, and Social Dynamics. BIOMAT 2022. Springer, Cham, DOI: 10.1007/978-3-031-33050-6_3, 2023. [27] Khachatryan, L., Xiang, Y., Ivanov, A. et al., Results and lessons learned from the sbv IMPROVER metagenomics diagnostics for inflammatory bowel disease challenge, Scientific Reports 13, 6303 DOI: 10.1038/s41598-023-33050-0, 2023. [26] M. Manzo, M. Giordano, L. Maddalena, iM.R. Guarracino, I. Granata, Novel Data Science Methodologies for Essential Genes Identification Based on Network Analysis, In Dzemyda, G., Bernataviciene, J., Kacprzyk, J. (eds), Data Science in Applications. Studies in Computational Intelligence, Vol. 1084, Springer, Cham., DOI: 10.1007/978-3-031-24453-7_7, 2023. [25] L. Maddalena, I. Granata, M. Giordano, M. Manzo, M.R. Guarracino, Integrating Different Data Modalities for the Classification of Alzheimer's Disease Stages, SN Computer Science, vol. 4, no. 249, DOI: 10.1007/s42979-023-01688-2, 2023. [24] I. Manipur, M. Giordano, M. Piccirillo, S. Parashuraman and L. Maddalena, Community Detection in Protein-Protein Interaction Networks and Applications, IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 20, no. 1, DOI: 10.1109/TCBB.2021.3138142, 2023. See also the Supplemental Material. [23] L. Maddalena, M. Giordano, M. Manzo, M.R. Guarracino, Whole-Graph Embedding and Adversarial Attacks for Life Sciences, in R.P. Mondaini (Ed), Trends in Biomathematics: Stability and Oscillations in Environmental, Social, and Biological Models: Selected Works from the BIOMAT Consortium Lectures, Rio de Janeiro, Brazil, 2021, Springer International Publishing, DOI: 10.1007/978-3-031-12515-7_1, 1--21, 2022. [22] I. Granata, I. Manipur, M. Giordano, L. Maddalena, M.R. Guarracino, TumorMet: A repository of tumor metabolic networks derived from context-specific Genome-Scale Metabolic Models, Scientific Data, DOI: 10.1038/s41597-022-01702-x, vol. 9, no. 607, 2022. [21] M. Giordano, L. Maddalena, M. Manzo, M.R. Guarracino, Adversarial attacks on graph-level embedding methods: a case study, Ann Math Artif Intell, DOI: 10.1007/s10472-022-09811-4, 2022. Free view-only version available here. [20] L. Maddalena, I. Granata, M. Giordano, M. Manzo, M.R. Guarracino, and for the Alzheimer's Disease Neuroimaging Initiative, Classifying Alzheimer's Disease Using MRIs and Transcriptomic Data, Proceedings of the 15th International Joint Conference on Biomedical Engineering Systems and Technologies - BIOIMAGING, ISBN 978-989-758-552-4, DOI: 10.5220/0010902900003123, pages 70-79, 2022. [19] I. Manipur, M. Manzo, I. Granata, M. Giordano, L. Maddalena, and M.R. Guarracino, Netpro2vec: a Graph Embedding Framework for Biomedical Applications, GNCS-INdAM Research Day on Computational Intelligence Methods for Digital Health, Vietri sul Mare (SA) and online, December 21, 2021. [18] M. Giordano, L. Maddalena, M. Manzo, and M.R. Guarracino, Adversarial Attacks on Graph Embedding: Applications in Computational Biology and Bioinformatics, Bioinformatics and Computational Biology Conference (BBCC 2021), online, December 1-3, 2021. [17] M. Giordano, L. Maddalena, M. Manzo, and M.R. Guarracino, Whole-Graph Embedding and Adversarial Attacks for Life Sciences, invited presentation at BIOMAT 2021 International Symposium, online, November 1-5, 2021. [16] M.R. Guarracino, M. Manzo, M. Giordano, L. Maddalena, On Resiliency and Robustness of Whole Graph Embedding, 11th International Conference on Network Analysis (NET 2021), online, October 18-20, 2021. [15] M. Giordano, I. Granata, M.R. Guarracino, L. Maddalena, M. Manzo, Graph Embedding for Biological Networks, Workshop How can Scientific Computing help to study Life Sciences?, Napoli and online, September 13, 2021. [14] M. Manzo, M. Giordano, L. Maddalena, and M.R. Guarracino, Whole Graph Embedding: Robustness and Vulnerability, Workshop How can Scientific Computing help to study Life Sciences?, Napoli and online, September 13, 2021. [13] M. Manzo, M. Giordano, L. Maddalena, and M.R. Guarracino, Performance Evaluation of Adversarial Attacks on Whole-Graph Embedding Models, In: D.E. Simos, P.M. Pardalos, and I.S. Kotsireas (Eds), Learning and Intelligent Optimization. Springer International Publishing, Lecture Notes in Computer Science vol. 12931, DOI: 10.1007/978-3-030-92121-7_19, ISBN: 978-3-030-92121-7, 219-236, 2021. [12] L. Maddalena, I. Manipur, M. Manzo, and M.R. Guarracino, On Whole-Graph Embedding Techniques, In: Mondaini R.P. (Ed) Trends in Biomathematics: Chaos and Control in Epidemics, Ecosystems, and Cells. BIOMAT 2020. Springer, Cham, DOI: 10.1007/978-3-030-73241-7_8, 115-131, 2021. [11] I. Manipur, M. Manzo, I. Granata, M. Giordano, L. Maddalena, and M.R. Guarracino, Netpro2vec: a Graph Embedding Framework for Biomedical Applications , in IEEE/ACM Transactions on Computational Biology and Bioinformatics, DOI: 10.1109/TCBB.2021.3078089, vol.19, no. 2, 2022. [10] L. Maddalena, I. Granata, I. Manipur, M. Manzo, and M.R. Guarracino, A Framework Based on Metabolic Networks and Biomedical Images Data to Discriminate Glioma Grades, in X. Ye et al. (Eds.), Biomedical Engineering Systems and Technologies. BIOSTEC 2020. CCIS 1400, Springer, ISBN: 978-3-030-72379-8, DOI: 10.1007/978-3-030-72379-8_9, 165--189, 2021. [9] I. Manipur, I. Granata, L. Maddalena, and M.R. Guarracino, Network Distances for Weighted Digraphs, in Y. Kochetov et al. (Eds.), Mathematical Optimization Theory and Operations Research, CCIS 1275, Springer Nature Switzerland, ISBN: 978-3-030-58657-7, DOI: 10.1007/978-3-030-58657-7_31, 389-408, 2020. [8] I. Manipur, I. Granata, L. Maddalena, and M.R. Guarracino, Clustering Analysis of Tumor Metabolic Networks, BMC Bioinformatics, ISSN: 1471-2105, DOI: 10.1186/s12859-020-03564-9, vol.21, n. 349, 2020. [7] I. Granata, M.R. Guarracino, L. Maddalena, I. Manipur, and P.M. Pardalos, On Network Similarities and Their Applications, in R.P. Mondaini (Ed.), Trends in Biomathematics: Modeling Cells, Flows, Epidemics, and the Environment: Selected Works from the BIOMAT Consortium Lectures, Szeged, Hungary, 2019, Springer International Publishing, 23-41, ISBN: 978-3-030-46306-9, DOI: 10.1007/978-3-030-46306-9_3, 2020. [6] L. Maddalena, I. Granata, I. Manipur, M. Manzo, and M.R. Guarracino, Glioma Grade Classification via Omics Imaging, in F. Soares, A. Fred, and H. Gamboa (Eds.), Proceed. 13th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC2020), Volume 2: BIOIMAGING, ISBN: 978-989-758-398-8, 82-92, 2020 (Best Paper Award). [5] I. Granata, M.R. Guarracino, V. Kalyagin, L. Maddalena, I. Manipur, and P. Pardalos, Model Simplification for Supervised Classification of Metabolic Networks, Annals of Mathematics and Artificial Intelligence 88, Springer, 91-104, 2020. [4] L. Antonelli, M. R. Guarracino, L. Maddalena, and M. Sangiovanni, Integrating Imaging and Omics Data: A Review, Biomedical Signal Processing and Control 52, 264–280, 2019. [3] I. Granata, M.R. Guarracino, V. Kalyagin, L. Maddalena, I. Manipur, and P. Pardalos, Supervised Classification of Metabolic Networks, 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 2688-2693, 2018. [2] I. Manipur, I. Granata, L. Maddalena, K.P. Tripathi, and M.R.Guarracino, Clustering Analysis of Tumor Metabolic Networks, Bioinformatics and Computational Biology Conference (BBCC2018), Naples, November 2018. [1] L. Antonelli, M. R. Guarracino, L. Maddalena, and M. Sangiovanni, Integrating Imaging and Omics Data: A Review, 17th International Symposium on Mathematical and Computational Biology - BIOMAT 2017, Moscow (Russia), November 2017.Last update: November 17, 2025
