Computational Data Science Laboratory (CDS-Lab), ICAR-CNR
Activities in Bio-Medical Imaging
SDI+: a Novel Algorithm for Segmenting Dermoscopic Images
- extracting preliminary information on possible confounding factors,
- accurately segmenting the lesion, and
- post-processing the result.
Performance evaluation of SDI+
Download the SDI+ Matlab code
How to cite the SDI+ software
References
SDI+ results
Experimental SDI+ results have been obtained on the test dataset for the ISIC 2017 lesion segmentation task [2], consisting of 600 dermoscopic images that include skin lesions of different types. Here you can find all the segmentation masks produced by SDI+. Some examples are given below.| A good result (JA=0.967757) | Image 16055 | 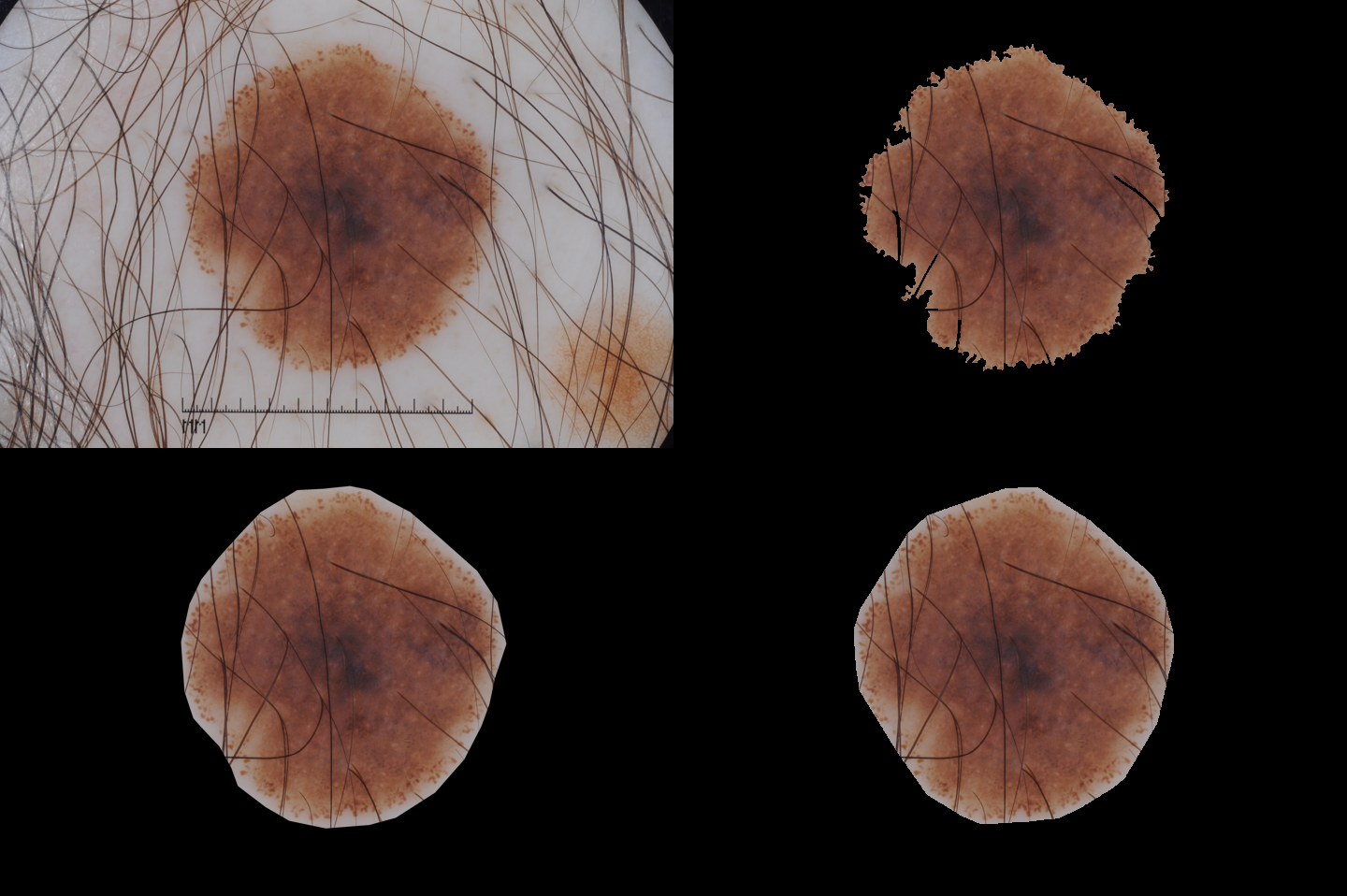 |
SDI+ initial segmentation |
| Ground truth | SDI+ final segmentation |
| A medium result (JA=0.608539) | Image 14219 | 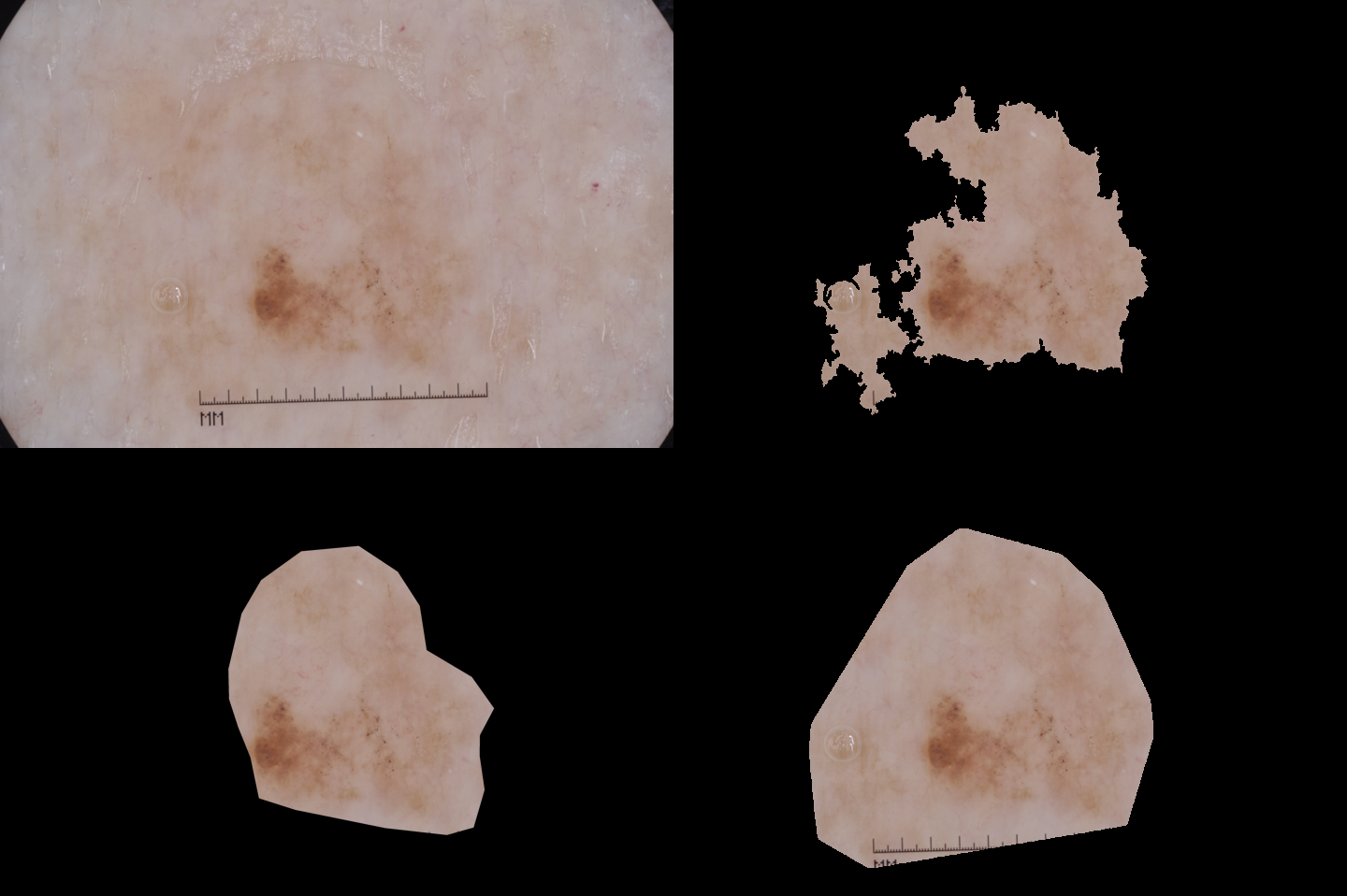 |
SDI+ initial segmentation |
| Ground truth | SDI+ final segmentation |
| One of the worse results (JA=0.205040) | Image 15645 | 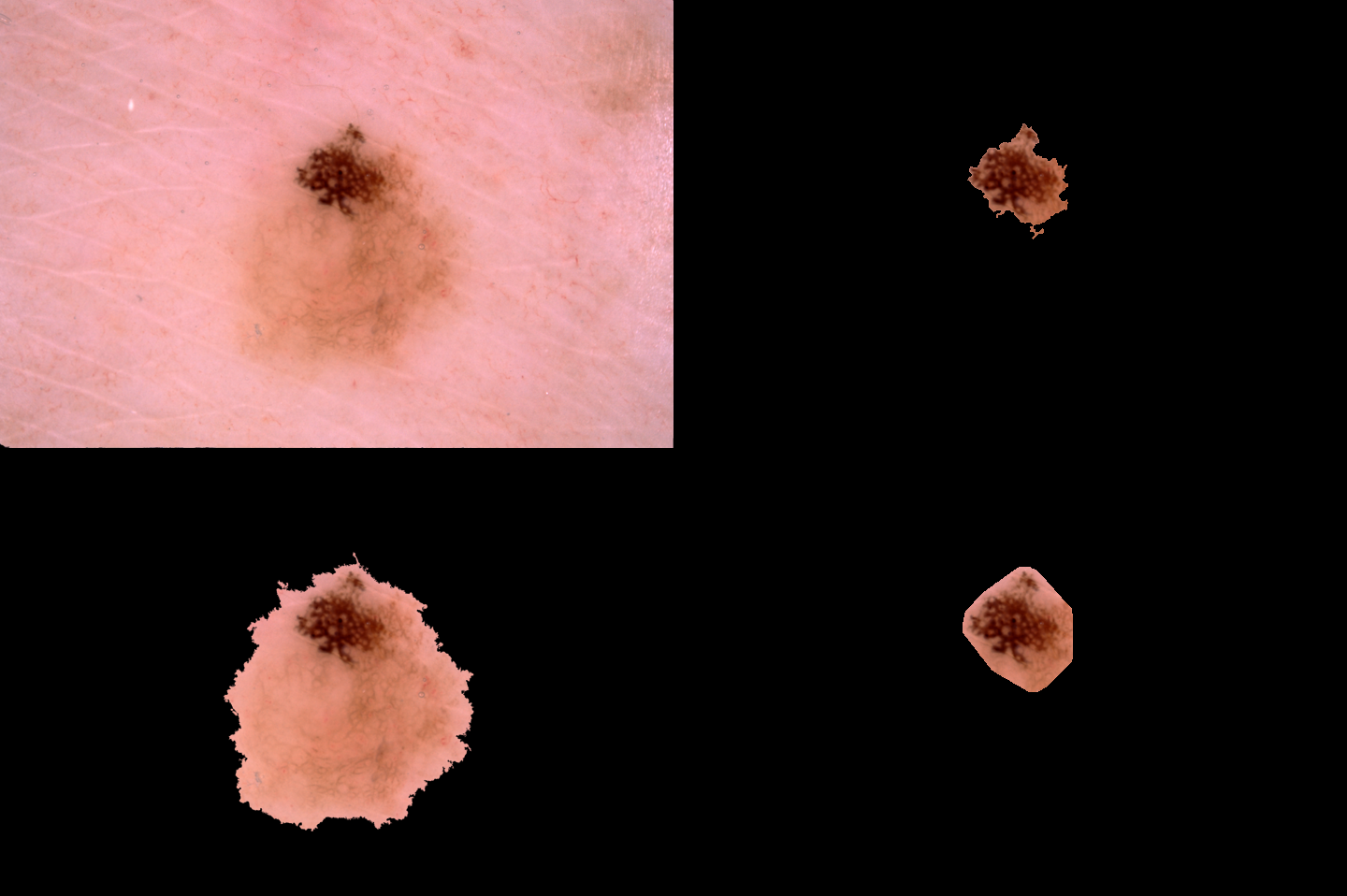 |
SDI+ initial segmentation |
| Ground truth | SDI+ final segmentation |
Performance evaluation of SDI+
The evaluation metrics adopted for the ISIC 2017 lesion segmentation task, described in [3], are| Pixel-level accuracy: | AC = (TP + TN)/(TP + FP + TN + FN); |
|---|---|
| Pixel-level sensitivity: | SE = TP/(TP + FN); |
| Pixel-level specificity: | SP = TN/(TN + FP); |
| Dice Coefficient: | DI = 2*TP/(2*TP + FN + FP); |
| Jaccard Index: | JA = TP/(TP + FN + FP); |
| AC | SE | SP | DI | JA | |
|---|---|---|---|---|---|
| SDI | 0.857 | 0.692 | 0.937 | 0.697 | 0.592 | SDI+ | 0.888 | 0.813 | 0.927 | 0.782 | 0.692 |
| AC | SE | SP | DI | JA | |
|---|---|---|---|---|---|
| Ambiguous (6) | 0.648 | 0.554 | 0.784 | 0.465 | 0.352 |
| Non-Uniform (31) | 0.772 | 0.501 | 0.936 | 0.490 | 0.388 |
| Clear (6) | 0.651 | 0.696 | 0.672 | 0.657 | 0.507 |
| Round (259) | 0.839 | 0.765 | 0.886 | 0.727 | 0.623 |
| Sharp (298) | 0.952 | 0.895 | 0.970 | 0.870 | 0.794 |
- Ambiguous (6 images): images whose GT is ambiguous in the shape it delineates;
- Non-Uniform (31 images): images that contain either more than one lesion or lesions having non-uniform color, easily perceived as multiple lesions;
- Clear (6 images): images that include lesions that are brighter than the surrounding skin;
- Round (259 images): images whose GT provides rough lesion contours, having roundness higher than 0.5;
- Sharp (298 images): images whose GT provides precise lesion contour, having roundness no higher than 0.5.
